This is an old revision of the document!
3D Modeling of Composite Membranes from TEM
🧬 Improving Protein Docking Accuracy with Iter-CONSRANK
KVL Staff on Project
Didier Barradas Bautista
didier.barradasbautista@kaust.edu.sa
Building 1, Level 0, Office 0125
![]() KAUST PI on Project
KAUST PI on Project
Luigi Cavallo
luigi.cavallo@kaust.edu.sa
Research Breakthrough
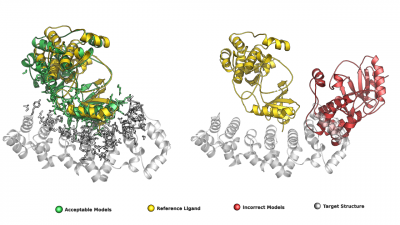
Protein–protein docking generates thousands of possible interaction models—but only a few are correct. In our latest collaborative work, we introduce Iter-CONSRANK, an iterative scoring algorithm that filters out incorrect models and enriches the ensemble with correct ones. 📊 Tested on two challenging datasets, Iter-CONSRANK increased the fraction of correct models by up to 8× for medium-difficulty targets and outperformed over 150 scoring functions in ranking accuracy. 🎯 The method is available for use in pre-processing docking ensembles or as an independent scoring tool. 🔬 This work was led in collaboration with Luigi Cavallo and our partners at the University of Naples “Parthenope”.
🔬 Research Visualization The stunning graphical abstract above showcases the innovative approach used in this groundbreaking research. The paper entitled “Impact of active layer morphology on salt permeability in RO composite membranes: 3D modelling from TEM geometry and effective membrane thickness” can be accessed here. Please note that all figures used in this article are directly from the original published paper.
🎯 KVL's Contribution KVL's visualization expert Dr. Ronell Sicat provided crucial support through:
- Advanced pre-processing of TEM scans
- Development and testing of segmentation methods
- Expert post-processing of segmentation data
- Creation of simulation-ready 3D geometries KVL also provided free access to Avizo software for data visualization, segmentation, and analysis and a powerful workstation with a pen-and-tablet interface that enabled faster manual segmentation of the TEM data. KVL's contribution is acknowledged in the paper.
📎 Read the full paper: [Link Placeholder]